
Databases
and Data Mining, Fall 2005
Lab Session 1
MSc Bio Informatics,
Peter van der Putten (putten-at-liacs.nl)
Ron Breukelaar (rbreukel-at-liacs.nl)
1.
Animal Trees
In this assignment we use a
data set of animals and their attributes. Using a decision tree classifier the
computer learns to classify animals into different categories (mammals, fish,
reptiles etc).
1.1 The data set can be found here. Without using
the data mining tool, draw a decision tree of three levels deep that classifies
animals into a mammal, bird, reptile, fish, amphibian, insect or invertebrate.
The decision tree doesn’t necessarily need to be correct for all animals.
Now we are going to let the
computer discover a decision tree itself. First download this zip file with
data sets to your desktop and unzip it. Open the zoo.arff data set in WEKA by
double clicking (or choose start
menu – weka – weka-3-4 – Weka Explorer – Open file).
1.2 What is the number of concepts, attributes and
instances in this data set?
Let us build some
classifiers. Go to the classifier tab. We will use 66% of the animals to build the
models, and the remaining 34% to evaluate the quality of the model., so select percentage split – 66%. First we will build a ‘naïve’ model that just
predicts the most occurring class in the data set for each animal. This
corresponds to a decision tree of depth 0. Click start to build a model.
1.3 What % of animals is correctly classified?
1.4 Into what category are all these animals classified
and why?
1.5 Now build a decision tree of depth 1 (a.k.a. a decision stump - select choose
– trees – decision stump). Draw the discovered decision tree.
1.6 What % of animals is correctly classified?
1.7 Give an example of an animal that would not be
classified correctly by this model.
1.8 Now build a decision tree of any depth (a.k.a. a J48 tree).
What % of animals is correctly classified?
1.9 Give an example of an animal that would not be
classified correctly by this model.
2.
Using pencil, paper and Naive Bayes for diabetes
diagnosis
In this exercise you will create
a Naive Bayes model on fictitious data for diagnosis of diabetes – without
using a mining tool. A real system could for instance be used to aid diagnosis,
or to identify patients at risk of becoming diabetic at a later stage.
2.1 Using the data below and the Naive Bayes algorithm, would
a young, slim female be classified as diabetic? With what probability?
2.2 Using the data below and the Naive Bayes algorithm,
would a old overweight male be classified as diabetic? With what probability?
2.3 Why is it
relatively straightforward to implement Naïve Bayes in a database?
|
GENDER |
AGE |
WEIGHT |
DIABETES |
|
F |
YOUNG |
|
NO |
|
F |
OLD |
|
NO |
|
M |
YOUNG |
|
NO |
|
M |
MIDDLE |
|
NO |
|
M |
OLD |
|
NO |
|
F |
YOUNG |
OVERWEIGHT |
NO |
|
F |
YOUNG |
SLIM |
NO |
|
F |
MIDDLE |
SLIM |
NO |
|
M |
YOUNG |
SLIM |
NO |
|
M |
MIDDLE |
SLIM |
NO |
|
M |
OLD |
SLIM |
NO |
|
F |
MIDDLE |
|
YES |
|
M |
OLD |
|
YES |
|
F |
MIDDLE |
OVERWEIGHT |
YES |
|
F |
OLD |
OVERWEIGHT |
YES |
|
M |
YOUNG |
OVERWEIGHT |
YES |
|
M |
OLD |
SLIM |
YES |
3.
Using the data mining tool for diabetes diagnosis
One of the data sets in zip
file is diabetes.arff. All patients in this data sets are females >21 yrs of
Pima Indian heritage from
3.1 Using WEKA Explorer create various classifiers to
fill in the table below. Try out various parameter settings to create better
models. Report on any non default settings used.
|
|
30% Hold Out Validation
accuracy |
10 Fold Cross Validation Average accuracy |
|
Decision Tree (J48) Non default settings: |
|
|
|
Naïve Bayes Non default settings: |
|
|
|
Nearest Neighbour (IBk) Non default settings: |
|
|
WEKA also offers a tool
(‘Experimenter’) to set up benchmarking experiments. For instance in the
example below Nearest Neighbor is benchmarked against Naïve Bayes on diabetes
and heart disease diagnosis data over 10 runs of 10 fold cross validation.
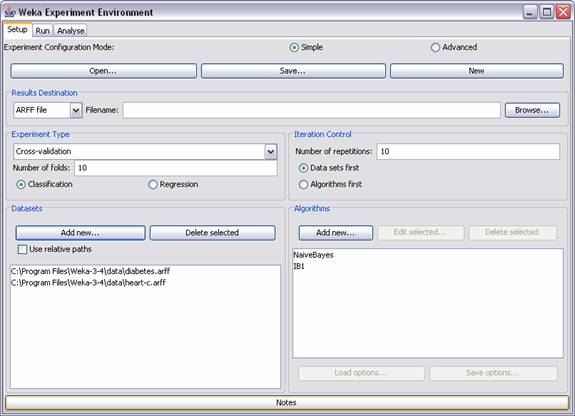
Repeat the experiments from
the previous exercise with 10 runs of 10 fold cross validation and note the
results below. Hints: hit New to
start; set up the experiment as in the picture above; hit run in the second tab; in the third tab select Scheme and Scheme options as Columns and select Perform test.
|
|
Average accuracy |
Standard Deviation |
|
Decision Tree (J48) Non default settings: |
|
|
|
Naïve Bayes Non default settings: |
|
|
|
Nearest Neighbor (IBk) Non default settings: |
|
|
4.
Classification for Bio Informatics Applications
(optional)
The zip file from assignment
1 contains a number of (bio)medical data sets. Most data sets contain a small
description in the header – to read this open the file in a text editor like
notepad. For a number of data sets (minimum 2) build a decision stump, a
decision tree and optionally another type of classifier. For each data set
note:
- The goal of the classifier: what needs to be
predicted
- A high level description of the attributes
available
- The classification accuracy for each classifier
type
- The patterns discovered by at least one of the
classifiers
- A suggestion of how such a prediction can be
used in practice